Research Focus
The group’s research focuses mainly on:
- Development of robust, generalizable neural networks (CNNs, Deep Learning)
- Data-/Annotation-efficient models based on Semi-/Self-Supervised Learning (SSL)
- Outlier-detection and imputation of incomplete data records
- Reconstruction of image and video data. e.g., by means of super-resolution
- Segmentation problems, particularly MRI Brain Segmentation
- Quantification of uncertainties in classification problems
- Development of interpretable features to improve user-/patient-communication
- Evaluation of algorithm performance and quantification of data-biases
- Translation of research results into industry and medical workflows
- Quantification of human anatomy based on image data (MRI, X-ray, CT) in the context of diseases such as dementia, tumor and traumas
Selected Research Projects
S. Doerrich, F. Di Salvo, J. Brockmann, C. Ledig, “Rethinking model prototyping through the MedMNIST+ dataset collection”, Scientific Reports, 15, 7669, 2025
The integration of deep learning based systems in clinical practice is often impeded by challenges rooted in limited and heterogeneous medical datasets. In addition, the field has increasingly prioritized marginal performance gains on a few, narrowly scoped benchmarks over clinical applicability, slowing down meaningful algorithmic progress. This trend often results in excessive fine-tuning of existing methods on selected datasets rather than fostering clinically relevant innovations. In response, this work introduces a comprehensive benchmark for the MedMNIST+ dataset collection, designed to diversify the evaluation landscape across several imaging modalities, anatomical regions, classification tasks and sample sizes. We systematically reassess commonly used Convolutional Neural Networks (CNNs) and Vision Transformer (ViT) architectures across distinct medical datasets, training methodologies, and input resolutions to validate and refine existing assumptions about model effectiveness and development. Our findings suggest that computationally efficient training schemes and modern foundation models offer viable alternatives to costly end-to-end training. Additionally, we observe that higher image resolutions do not consistently improve performance beyond a certain threshold. This highlights the potential benefits of using lower resolutions, particularly in prototyping stages, to reduce computational demands without sacrificing accuracy. Notably, our analysis reaffirms the competitiveness of CNNs compared to ViTs, emphasizing the importance of comprehending the intrinsic capabilities of different architectures. Finally, by establishing a standardized evaluation framework, we aim to enhance transparency, reproducibility, and comparability within the MedMNIST+ dataset collection as well as future research.
Authors: Sebastian Doerrich, Francesco Di Salvo, Julius Brockmann, Christian Ledig
[Preprint], [Publication], [Code], [Benchmark], [BibTeX](612.0 B)
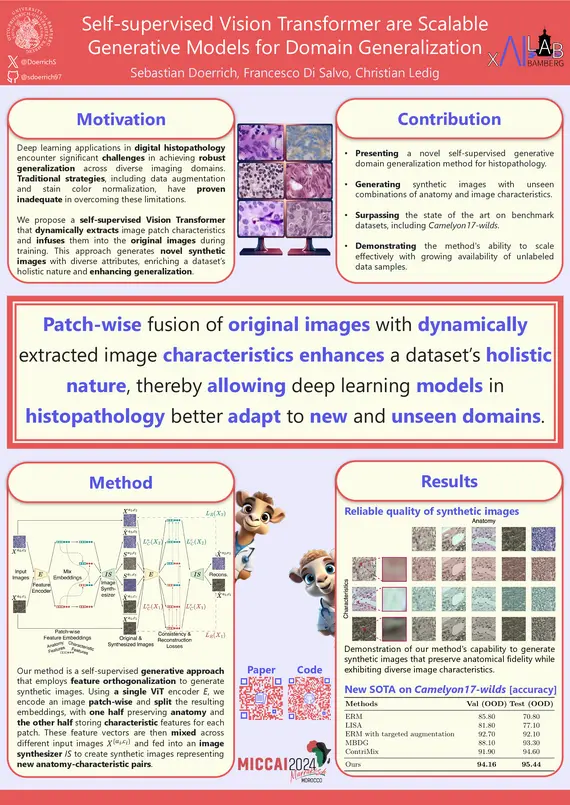
S. Doerrich, F. Di Salvo, C. Ledig, "Self-supervised Vision Transformer are Scalable Generative Models for Domain Generalization", MICCAI, 2024
Despite notable advancements, the integration of deep learning (DL) techniques into impactful clinical applications, particularly in the realm of digital histopathology, has been hindered by challenges associated with achieving robust generalization across diverse imaging domains and characteristics. Traditional mitigation strategies in this field such as data augmentation and stain color normalization have proven insufficient in addressing this limitation, necessitating the exploration of alternative methodologies. To this end, we propose a novel generative method for domain generalization in histopathology images. Our method employs a generative, self-supervised Vision Transformer to dynamically extract characteristics of image patches and seamlessly infuse them into the original images, thereby creating novel, synthetic images with diverse attributes. By enriching the dataset with such synthesized images, we aim to enhance its holistic nature, facilitating improved generalization of DL models to unseen domains. Extensive experiments conducted on two distinct histopathology datasets demonstrate the effectiveness of our proposed approach, outperforming the state of the art substantially, on the Camelyon17-wilds challenge dataset (+2%) and on a second epithelium-stroma dataset (+26%). Furthermore, we emphasize our method’s ability to readily scale with increasingly available unlabeled data samples and more complex, higher parametric architectures.
Authors: Sebastian Doerrich, Francesco Di Salvo, Christian Ledig
[Preprint], [Publication], [Code], [BibTeX](612.0 B)
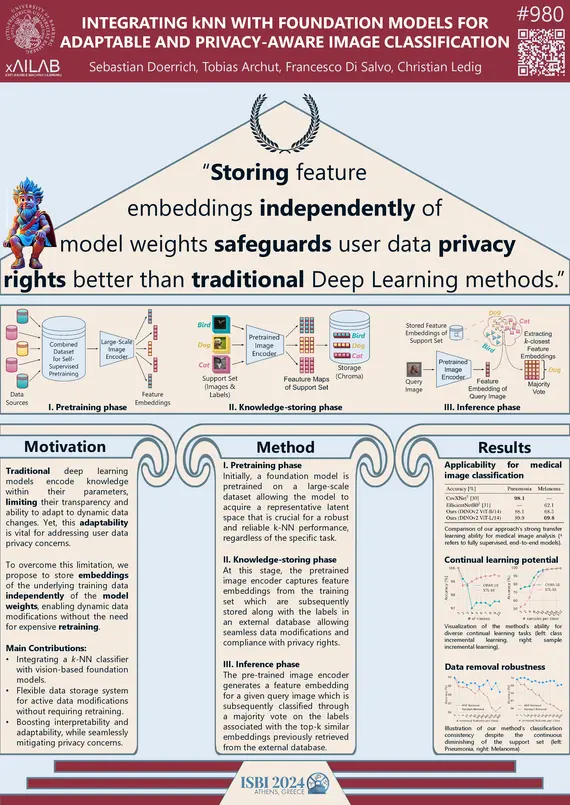
S. Doerrich, T. Archut, F. Di Salvo, C. Ledig, "Integrating kNN with Foundation Models for Adaptable and Privacy-Aware Image Classification", IEEE ISBI, 2024
Traditional deep learning models implicity encode knowledge limiting their transparency and ability to adapt to data changes. Yet, this adaptability is vital for addressing user data privacy concerns. We address this limitation by storing embeddings of the underlying training data independently of the model weights, enabling dynamic data modifications without retraining. Specifically, our approach integrates the k-Nearest Neighbor (k-NN) classifier with a vision-based foundation model, pre-trained self-supervised on natural images, enhancing interpretability and adaptability. We share open-source implementations of a previously unpublished baseline method as well as our performance-improving contributions. Quantitative experiments confirm improved classification across established benchmark datasets and the method’s applicability to distinct medical image classification tasks. Additionally, we assess the method’s robustness in continual learning and data removal scenarios. The approach exhibits great promise for bridging the gap between foundation models’ performance and challenges tied to data privacy.
Authors: Sebastian Doerrich, Tobias Archut, Francesco Di Salvo, Christian Ledig
[Preprint], [Publication], [Code], [BibTeX](612.0 B)